Donor Screening for Transfusion-Transmitted Infections (TTI)
Screening donated blood for transfusion-transmitted infections is critical to ensure the safety of blood and blood components. Nucleic acid technology (NAT) testing, when used in combination with immunoassays, can detect infections and help reduce the risk of viruses entering the blood supply.
NAT for Blood Donor Screening
The importance of blood screening was highlighted in the 1980s when thousands of patients with hemophilia were infected with hepatitis C virus (HCV) and HIV as a result of receiving contaminated blood products. At the time, testing of blood products by NAT (a highly sensitive molecular technique) revealed high HCV contamination rates, and promoted the idea of using NAT in addition to antigen and antibody screening.
Various markers of infection appear at different times after infection. The first detectable target to appear is nucleic acid, which is part of the native infectious agent, followed a few days later by antigens and subsequently antibodies that develop during an immune response. NAT, which detects the presence of a virus even in very small amounts, can detect infections earlier than immunoassays (used to detect antigens and antibodies). Thus, NAT used in combination with immunoassays can further reduce the risk of viruses entering the blood supply.
What is NAT?
Most NAT assays involve a nucleic acid extraction step followed by amplification and detection/quantitation. NAT includes a variety of methods, such as polymerase chain reaction (PCR), reverse transcription polymerase chain reaction (RT-PCR), transcription-mediated amplification (TMA), ligase chain reaction (LCR), strand displacement amplification (SDA), and nucleic acid sequence-based amplification (NASBA).
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are the two main types of nucleic acid. They are polymers made up of nucleotides consisting of a pentose (5 carbon) sugar, a phosphate group, and a nitrogen-containing base. There are five different bases that bind together to form specific complementary pairs: cytosine binds to guanine, and adenine binds to thymine in DNA and uracil in RNA. Oligonucleotides are man-made polymers that are single-stranded sequences consisting of at least two nucleotides (usually about 20-40 nucleotides in length). They are specifically designed to bind complementarily to a conserved region of the target sequence. These regions are chosen because they are not easily mutated.
What is TMA?
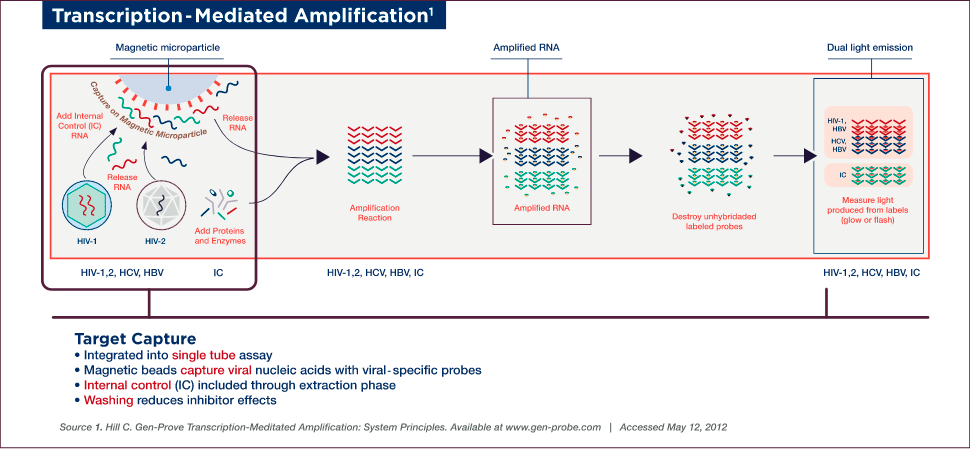
TMA (transcription-mediated amplification) is a single-tube, isothermal (conducted at a constant temperature) NAT assay that allows simultaneous detection of multiple viruses. This NAT assay consists of three steps (target capture, amplification, and detection). TMA uses three different types of oligonucleotides: capture oligonucleotides in the target capture step, the primers in the amplification step, and probes in the detection step.
TMA Target Capture
Target capture, the first step of the assay, involves capturing viral nucleic acids with viral-specific oligonucleotides linked to magnetic beads. Samples are prepared for testing by lysing viruses to release the genetic material — no pretreatment or handling is required. An internal control (IC) is also introduced to each reaction tube during the target capture step, which controls for proper reagent addition and performance of each step throughout the assay procedure. The IC is set at a low target level to detect variations in amplification efficiency due to sample, reagent, or procedural problems.
Target capture is accomplished using capture oligonucleotides that have two sections, a poly-dA tail (string of deoxyadenosine [adenine plus deoxyribose pentose sugar] residues) and a section homologous to highly conserved regions of viral genomic material, which hybridizes to the target. The hybridized target and IC are captured onto magnetic microparticles that have poly-dT (poly-deoxythymidine) molecules covalently attached. Unbound material is washed away to remove non-specific material and to minimize potential inhibitors.
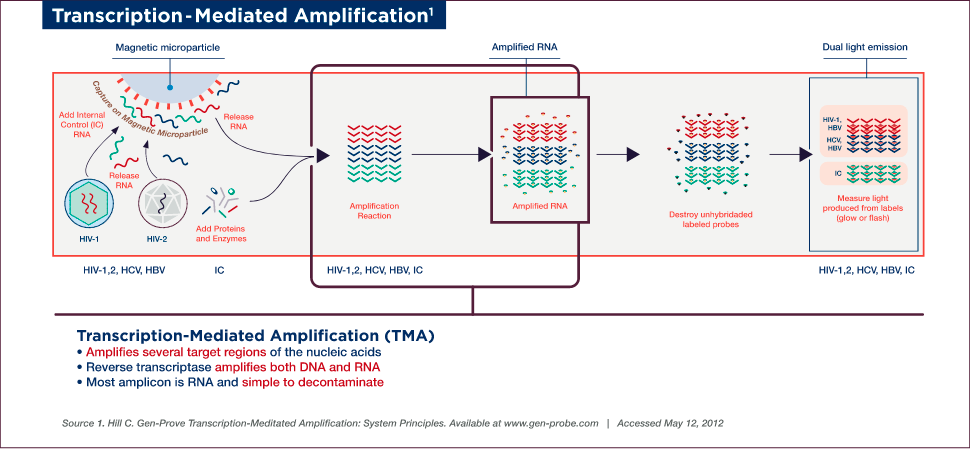
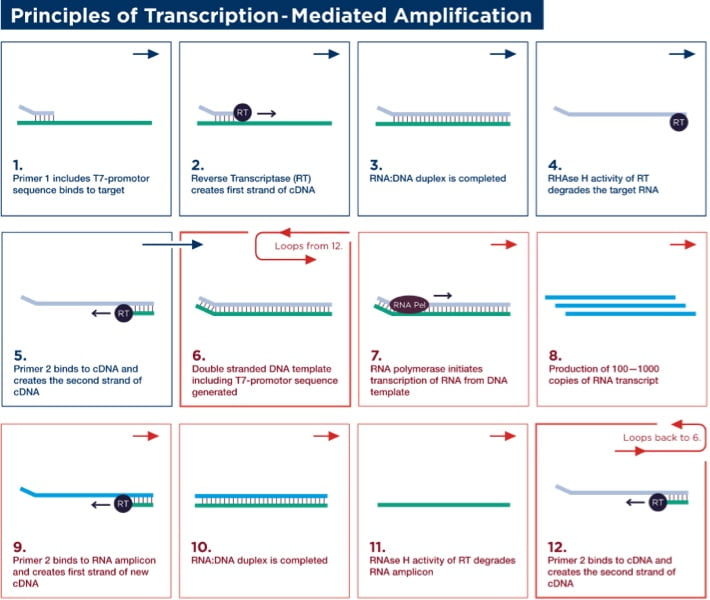
Target Amplification
Target amplification occurs via TMA, which utilizes two enzymes, a reverse transcriptase (with RNA-dependent and DNA-dependent DNA polymerase activities, as well as RNase H activity) and bacteriophage T7 RNA polymerase. Primer 1 (which includes a T7-promoter sequence) binds to the target and is extended by the reverse transcriptase to create the first strand of complementary DNA (cDNA). Once the RNA/DNA duplex is completed, the RNase H activity of reverse transcriptase digests the RNA strand from the RNA/DNA duplex, leaving only the cDNA. Primer 2 (non-T7 primer) then binds to the cDNA and the reverse transcriptase catalyzes synthesis of another new DNA strand using the cDNA as a template, thereby generating the double-stranded DNA intermediate, which includes the T7-promoter sequence. The T7 RNA polymerase now recognizes the promoter sequence on the DNA intermediate and transcribes multiple (100–1000) copies of RNA amplicon. The RNA amplicons created bind to primer 2 and are extended by reverse transcriptase to create the first strand of new cDNA, thus generating an RNA/DNA hybrid duplex to repeat the cycle and resulting in approximately 1 billion-fold amplification in 30 minutes.
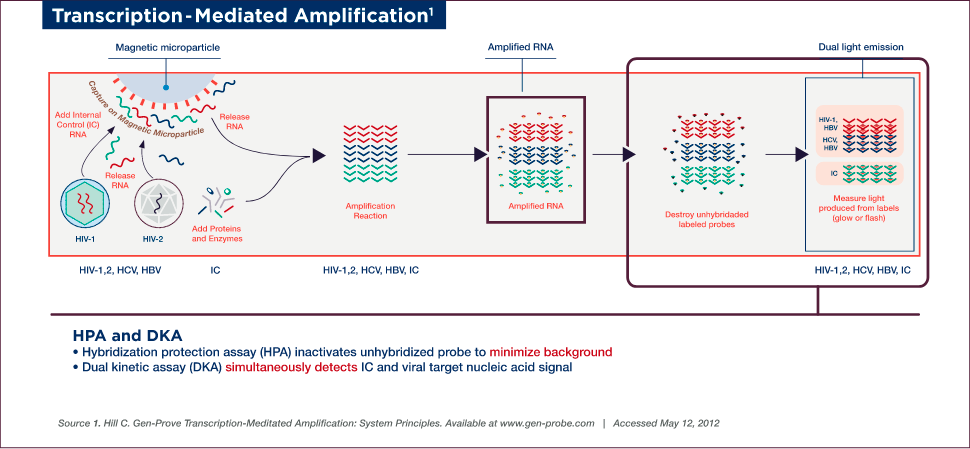
TMA Detection
Detection is achieved by the hybridization protection assay (HPA), which uses single-stranded, acridinium ester (AE)-labelled nucleic acid probes that are complementary to the amplicon. These probes hybridize specifically to the amplicon, while the AE label on any unhybridized probe is inactivated by hydrolysis during the selection step following hybridization.
A dual kinetic assay (DKA) is used to detect the viral nucleic acid and IC signals simultaneously. The assay employs single-stranded probes labelled with two different types of AE molecules, which differ in the kinetics of light emission, thereby allowing for the detection of two analytes in a single reaction. The amplicon specific to viral nucleic acid is detected using probes with relatively slower kinetics of light emission (glower signal), while the IC-specific amplicon is detected using a probe with rapid emission of light (flasher signal). The chemiluminescent signal produced by the hybridized probes are measured in a luminometer and reported as Relative Light Units (RLU).
Advantages of TMA for Detecting Transfusion-Transmitted Infections
TMA has several advantages compared with conventional PCR-based assays:
- While PCR requires thermal cycling instrumentation, TMA requires much more modest temperature control for isothermal amplification. The three steps of the TMA assay (target capture, amplification, and detection) are conducted in a single tube, with fewer mechanical steps and faster processing time than PCR.
- The risk of cross-contamination (a major concern in PCR labs) in the TMA assay is very low, as it uses a single-tube reaction with few processing steps.
- The lack of a separate step between purification and amplification simplifies instrumentation and processing.